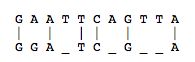Biology and Computers: A lesson in what is possible
History and Impact of Bioinformatics on Biology
Disclaimer: “I’m a student in the History of Computing class at San Jose State University ( http://www.cs.sjsu.edu/~mak/CS185C/ ). This is a work in progress that will turn into a final article by the end of the semester. I welcome your comments and advice!”
This article will deal with how computation has changed the face of biology and created a new field in Bioinformatics. I want to go over problems that biologists faced and how these problems were solved through the use of computers. Additionally, I would like to cover a history of Bioinformatics, beginning with algorithms like the Needleman-Wunsch, and ending with the tools available today and the current state of the Human Genome Project (which is not done yet)
Introduction
The 20th century saw major advances in biology, with the discovery of DNA and greater understanding of the underlying mechanisms powering life functions. With these discoveries came great challenges, some that couldn’t be solved in the lab by an experiment. For example, the DNA that makes up the human genome contains over three billion base units, only a few segments of which are actually genes. Parsing, searching, and organizing the three billion letters of human DNA are problems that computers are uniquely suited to handle. But when DNA was being studied, computer development was in its infancy. When scientists were ready to begin breaking apart human DNA in the late 1980's, the picture had changed. Computers at that point were well on their way to becoming ubiquitous and scientist were in a position to use them towards achieving their grand vision.
There are other interesting issues that pop up in biology which require processing power. In chemistry, bond angles of molecules are studied in order to better understand the dynamics of reactions. On a small scale, this is quite manageable. However, proteins are composed of hundreds, if not thousands of atoms, with an equal or greater amount of bonds. Figuring out how bond angles will affect the folding of a protein becomes a difficult if not insurmountable task. A computer, on the other hand, can easily keep track of millions of bond angles.
Much of Biology is focused on creating new therapies to heal the sick and improve the quality of life of those who have chronic conditions. Here too, computers are quickly changing the picture to make a significant impact. From changing the way that diabetics dose their insulin, to revolutionizing drug testing, computer scientists are poised to have as significant of an impact as and doctor or scientist.
Brief Definition of Bioinformatics
Bioinformatics is a scientific field that uses the tools of Computer Science and Information Technology towards achieving a better understanding of Biology. [4]
Databases are commonly used to house information about DNA and RNA sequences, such as their species of origin, who discovered the sequence, what the actual sequence looks like, what variants there might be. There are several large databases. The National Center for Biotechnology Information hosts GenBank, a collection of all publicly available DNA sequences. Swiss-Prot is a Protein information database hosted by the Swiss Institute for Bioinformatics.
In addition to Databases, many bioinformatics sites also contain tools for analyzing sequences. The NCBI has a set of tools collectively called BLAST, which stands for Basic Local Alignment Search Tool. A nucleotide BLAST will take a user provided nucleotide sequence and run it against an NCBI species-specific database to search for matches using a sequence alignment algorithm. This kind of information can be invaluable for elucidating the function of the submitted sequence. If the sequence is human and aligns with a mouse sequence that corresponds to a gene for determining eye-color, it’s possible the human sequence might have a similar function.
Tools, however, are not limited to sequence comparisons and database searches. Programs like Rosetta@home help get at solving the problem of protein folding mentioned above, by using algorithms to predict how a given sequence of amino acids might fold into a protein.
The Human Genome Project
At the beginning of the 1990’s there was an ambitious undertaking to document the entire human genome. A genome contains the hereditary information for a species. It is composed of DNA and is held in the chromosomes of cells.[10]
Quick Biology Primer
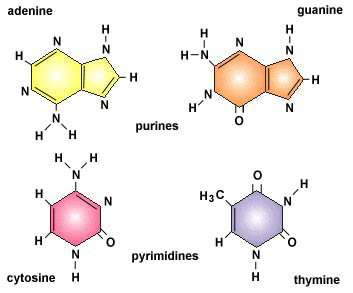
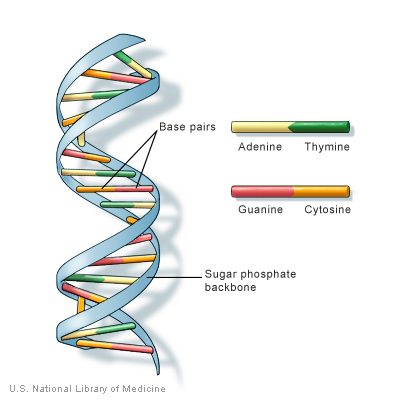
DNA is the basic construction unit for biological organisms. It is composed of a collection of molecules, termed nucleotides, arranged in a predetermined fashion on a phosphate scaffold. The four nucleotides are Adenine, Guanine, Cytosine, and Thymine. They are represented with the following letters, respectively: A, G, C, and T. With these letter designations, a DNA strand can be visualized as a string of nucleotide letters, e.g., AGTAACGGTTTAGAGTGA.
 Source
Source
For every sequence of DNA, there is a complimentary sequence that goes with the original strand. Together, they form a double strand, or double helix. The complimentary strand is composed of nucleotides complimentary to the sequences of the original strand. The pairings are as follows: A-T, G-C, C-G, and T-A. Thus, the following sequence, AGGATCA, would have the following complimentary strand, TCCTAGT. For more information on DNA and Nucleotides, see here.
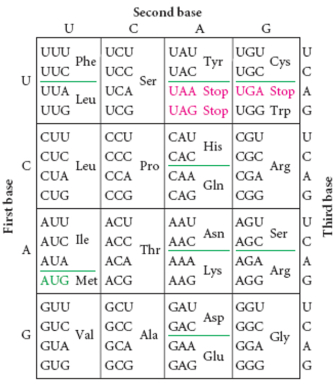
From the DNA blueprint, cells make RNA, which is then rearranged into three-member groupings called Amino Acids. RNA looks much like DNA only Thymine is replace with Uracil, represented by U. The corresponding nucleotide is the same: A-U, U-A. For more information on RNA, see here.
Amino acids are the building blocks for proteins, which go on to perform a number of tasks, from forming structures like cell walls or the nucleus to regulating processes such as blood coagulation and antigen detection. Amino acids form in predictable patterns and are also represented by a set of letters. (show image here).
The Human Genome Project involved closely coordinated work amongst many groups all around the world. In order to facilitate cooperation amongst all of the scientists and coordinate efforts, data was compiled on online databases before being published.[9] This was one of the first major uses of Bioinformatics. The relational databases that were created for the project were large and complex, since they contained not just sequences, but a great deal of information regarding each particular sequence if it pertained to a known gene. Three of the major databases that were formed were the NCBI Genbank, USCS Genome Browser, and the Sanger Institute’s Ensembl database.
The utility of the current databases extends beyond simply supplying discovered sequences. It also provides tools for exploring new things about each sequence. For example, sequence alignment allows one to discover how similar one sequence is to another. This can be useful for determining the function of a particular gene sequence. For example, if you have the sequence for a gene that determines eye color in chimpanzees, you can compare the sequence against the human genome to see if you can find a similar sequence. This is accomplished by using sequence alignment software tools that incorporate powerful algorithms. Two of the best-known algorithms are Needleman-Wunsch and Smith-Waterman.
Needleman-Wunsch
Originally presented in a 1970 in an article in the Journal of Molecular Biology, the Needleman-Wunsch algorithm presented a way to perform comparisons on two sequences of letters, showing which segments of those sequences were “conserved.”[5] The Needleman-Wunsch algorithm is an example of a global sequence alignment. A global sequence alignment takes two very similar sequences and attempts to find the optimal way to line up letters in each sequence while minimizing the amount of gaps it creates in a sequence. Needleman-Wunsch accomplishes this through two sets of rules.
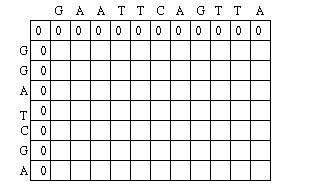
First, it applies a scoring scheme matrix for aligning the sequences. The matrix is created by filling each position in the two dimensional matrix with the score for a different pairing(insert picture here). This scoring scheme punishes mismatches, and importantly, punishes more severely for certain types of mismatches. It does this by taking into consideration fundamental aspects of nucleotides and amino acids that make them unique. For example, nucleotides can be separated into two chemically different subgroups: pyrimidines and purines. T and C represent pyrimidines, and A and G represent the chemically larger purines. Amino acids can also be grouped into chemically similar subgroups. The idea is to score chemically dissimilar nucleotides and amino acids more severely based on the degrees of difference.[5]
The second set of rules pertains to the creation of gaps. The goal is to avoid gaps if possible by assigning a large penalty to the creation of a gap. If a gap is created, extending the gap is not punished, but creating another gap nearby is punished even more severely. The goal is to minimize the amount of gaps, and to maximize the size of gaps.
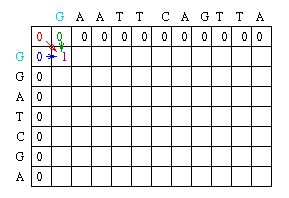
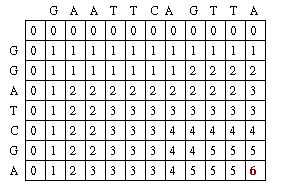
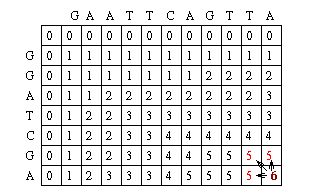
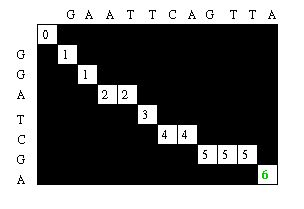
The Needleman-Wunsch keeps track of the best sequence in a second matrix called the traceback matrix. The traceback matrix keeps track of what path needs to be taken through the matrix in order to build the optimal sequence. It does this by noting the score for each position in the matrix.[5]
Starting at the farthest diagonal end of the matrix, the sequence is created by following the highest scoring path. If the movement is diagonal, it means the two sequences are aligned, whereas an upwards or sideways move represents a gap creation. [6]
The Needleman-Wunsch algorithm is most useful when comparing two sequences of similar length with a large degree of similarity. For sequences of starkly different lengths, local sequence alignment is more appropriate. Smith-Waterman is widely used to perform local sequence alignment.[1]
The Smith-Waterman algorithm functions very similarly to the Needleman-Wunsch algorithm with a crucial difference: the Smith-Waterman algorithm never penalizes a pairing below 0. In other words, no negative scores are generated on the matrix. As a result, the matrix may only have a small portion with scores filled. This filled portion represents the local alignment.
BLAST, the popular tool for aligning sequences on the NCBI website implements the Smith-Waterman algorithm. The Smith-Waterman algorithm is particularly suited for database searching since it is better than the Needleman-Wunsch algorithm at finding short similar sequences across large databases of different genomes.[1]
Foldit: Folding Proteins at Home
Protein folding is one of the big challenges facing biology and medicine. When amino acids are joined together to form proteins, they don’t remain a long chain. Instead, the chain folds to form a stable structure. The design of this structure is paramount: proteins are only meant to interact in limited ways in the body. A change in the folding pattern of a protein might completely change its function or render it inactive.
Misfolded proteins can often lead to disease. In sickle cell anemia, a single changed nucleotide in the genome leads to a hemoglobin (red blood cell) protein with an amino acid composition that is different by a single amino acid. This small difference results in a misfolded hemoglobin protein that fundamentally changes the shape of red blood cells. Analysis of the structure of hemoglobin through the use of protein structure software makes the mutation obvious.[11]
Viruses and bacteria have several types of proteins that make up their outer shells. Determining the structure of these proteins can provide scientists and doctors with a way to attack the disease. For this, protein structure software is also crucial. At the University of Washington, Professor David Baker utilizes software called Rosetta@home in order to help determine protein structures. Rosetta is a “crowd-sourced” program. It is made available to the public so that an individual can install it and run it in the background while the computer is idle. By doing this, Rosetta is able to use a “pseudo” supercomputer in the form of this massive network to perform its many calculations.[8]
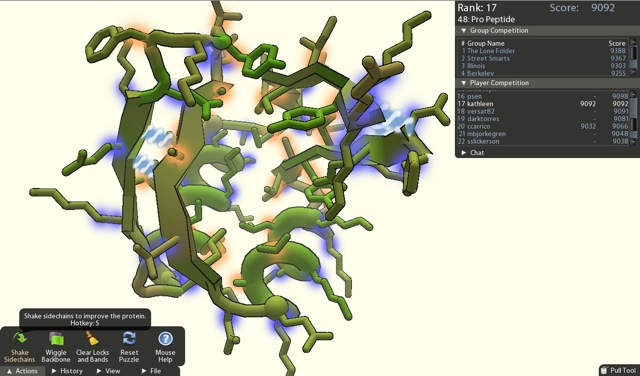
Another program built by the Baker lab is even more interesting. Foldit is a game designed to allow members of the public to toy with protein confirmations in order to arrive at an “ideal” confirmation. Players are trained through the games tutorials to learn how they can manipulate the bonds of a proteins many molecular components. Poor confirmations are formed when the bond angles of molecular components bring atoms too close to each other. This results in a high energy confirmation and is unfavorable and unlikely to exist in nature. The goal of the game is for players to play with this bond angles until they can lower the energy of level of the entire protein to the lowest possible level. Players compete with one another to see who can achieve the lowest energy and, thus, the highest score.[7]
This formed the foundation for David Baker’s experiment: to see if the collected brain power of many game players could outperform the Rosetta program. The result was spectacular. Not only were the players able to beat the Rosetta program, they were also able to arrive at the actual protein confirmation for a protein that scientists have been attempting to discover for over 10 years! This particular protein is considered an to be an important target for the fight against HIV. The correct confirmation was buffered by experimental evidence of its correctness.[3]
This achievement was made possible because of the way that players approached the problem differently from the way Rosetta did. Rosetta attempted to find the correct protein by carefully moving through the protein, repositioning bonds while maintaining the lowest possible energy at all times. If the energy levels rose to high, Rosetta would abandon that path and move on to another. Players, on the other hand, would accept high energy levels if they could foresee eventually lowering them later on.[3]
This was a fundamental difference. Rosetta was unable to plan so far ahead to see the value in accepting high energy states to begin with. Given that this is only the beginning and that the software continues to improve, it will be exciting to see how profound of an impact this computer software can have on future therapeutic development.
The Future
In addition to both sequencing and protein structuring, there are many ways in which computer science will continue to have a profound impact on biology and medicine. Two ways in which computer science will affect medicine in the near future stand out: diabetes and therapeutic drug testing.
Diabetes is a fast growing health crisis in the United States and other countries. There are currently many treatment options available for those with type 2 diabetes, and it's late onset means that some complications of diabetes never develop. Those with type 1 diabetes, on the other hand, the disease typically presents early and as a result, there is more time for diabetes complications to arise. The next generation of diabetes is aimed at helping this population.
The artificial pancreas is a concept being pursued by several companies and organizations. The idea behind the artificial pancreas is to provide the diabetes patient with a closed loop system: a computer monitor that measure blood glucose levels in real time, an attached insulin pump to bring glucose levels down, a glucagon pump to provide emergency glucose when levels are dangerously low, and a robust software package to bring it all together.[2]
Individually, all of these components are available, except for a full software automation system. This is a future computer science problem: developing accurate algorithms to determine when the patient should get insulin, and when they should get glucose.[2]
Therapeutic drug testing is also poised for fundamental change with continued computerization. For much of recorded history, humans have tested medicine on animals before using them on the sick. The reason for this is obvious, but there are relevant concerns that challenge the status quo. Setting aside the valid ethical concerns, there remains the fact that while sequence analysis can find the similarity among species that provide the rational for animal drug models, any given non-human animal will never be able to serve as a 1:1 mimic for the human body. Differences genome will always create variability that can potentially lead to problems when a drug is eventually introduced into humans.
On way to deal with this problem is to build computerized human models that can simulate the way that the body will react to a therapy. As scientists discover more and more about the human body, more and more information can be fed into the human model to make it more accurate. It’s not hard to imagine that the point might eventually be reached where the computerized human model might be more accurate than any animal model. This would have the peripheral benefit of considerably lowering drug costs, since the most expensive part of making a drug is not the creation of the drug, but the testing.
As computer science continues to develop, it is easy to see that it will become more and more integrated with the disciplines which are able to make use of it. One discipline that has and will continue to benefit from computerization is Biology.
References
1. Cold Spring Harbor Laboratory Press. (2004). Examples of global and local alignments of two proteins. Retrieved November 23, 2011, from bioinformaticsonline.org: http://www.bioinformaticsonline.org/ch/ch03/supp-1.html
2. Juvenile Diabetes Research Foundation International. (2010). Algorithm. Retrieved December 08, 2011, from JDRF Web Site: http://www.artificialpancreasproject.com/about/algorithm.html
3. Khatib, F., Baker, D., & al., e. (2011, September 18). Crystal structure of a monomeric retroviral protease solved by protein folding game players . Nature Structural & Molecular Biology .
4. NCBI. (2004, March 29). Primer. Retrieved October 10, 2011, from NCBI.gov: http://www.ncbi.nlm.nih.gov/About/primer/bioinformatics.html
5. Needleman, S., & Wunsch, C. (1970). A general method applicable to the search for similarities in the amino acid sequence of two proteins. Journal of Molecular Biology , 443-453.
6. Rouchka, E. C. (2001). Needleman- Dynamic Programming Tutorial. Retrieved November 18, 2011, from Eric Rouchka Dynamic Programming Web Site: http://www.avatar.se/molbioinfo2001/dynprog/dynamic.html
7. University of Washington. (2011, November). Foldit Solve Puzzles for Science. Retrieved December 02, 2011, from Foldit Web Site: http://fold.it/portal/info/science
8. University of Washington. (2010, November 2010). Rosetta@home. Retrieved December 02, 2011, from Rosetta@home Web site: http://boinc.bakerlab.org/rosetta/
9. Wellcome Trust Sanger Institute. (2011, May 03). History. Retrieved November 18, 2011, from Wellcome Trust Sanger Institute Web Site: http://www.sanger.ac.uk/about/history/hgp/#tabs-3
10. Wikipedia. (2011, November 1). Genome. Retrieved November 15, 2011, from Wikipedia.org: http://en.wikipedia.org/wiki/Genome
11. Zvelebil, M. J., & Baum, J. O. (2008). understanding bioinformatics. New York, NY: Garland Science.